While GM crops of past years contain one to few traits, the continuous discovery of new trait genes would mean that over time, crops could end up with a large number of transgene insertions. If they were dispersed throughout the genome, extensive breeding would be needed to reassemble all of them into a single breeding line. Stacking new DNA to a preexisting transgenic locus insures that the package of transgenes can be transmitted through breeding programs as a single locus rather than segregating loci.
The former study from Dr. David W. Ow ’s research group in South China Botanical Garden of Chinese Academy of Sciences reported an in planta gene stacking method using the Bxb1 integrase for site-specific integration followed by the Cre recombinase for removal of unneeded DNA (HOU et al., 2014 Molecular Plant 7:1756-1765). This method permits the sequential addition of transgenes as each integrating molecule brings a new recombination target for the next round of integration.
However, a need arises later on that may require removal of existing transgenic DNA. To accomplish this goal, Dr. David W. Ow’s research group developed a method about deleting or replacing preexisting transgenes by the same recombinase-mediated gene stacking systems as described in the former study mentioned above .
Microparticle bombardment was used to deliver pHL002 (Fig. b) along with a Bxb1 integrase-expressing construct into explants of line 23.C.4-9.d8.BC1 (Fig. a), which was created by stacking two transgenes in a tobacco target line. Bxb1-mediated site-specific integration of pHL002 inserts OsO3L2-2B into the target locus placing directly oriented lox sites to flank the previously stacked transgenes gus, luc and gfp (Fig. c).
This should permit the subsequent deletion of the previously stacked transgenes to produce the configurations shown in Figure 1d or e. Moreover, the new structure with a single attachment site (attB or attP) can continue to permit subsequent rounds of gene stacking or gene replacement. Whether this is used for deleting preexisting transgenes, replacing with new versions of the same gene or replacing with a completely new transgene, this transgene locus editing feature extends the flexibility of the Bxb1/Cre recombinase-mediated system for transgene stacking.
As opposed to patent licenses needed for use of sequence-specific nuclease-based tools, using this gene deletion/replacement strategy has freedom to operate and is more applicable in crop improvement.
Postdoctoral fellow CHEN Weiqiang and former Ph.D. student Gurminder Kaur were the co-first authors, Dr. David W. Ow was the corresponding author.
The relevant paper entitled “Replacement of stacked transgenes in planta” was published in Plant Biotechnology Journal. For further reading, please refer to:https://onlinelibrary.wiley.com/doi/pdf/10.1111/pbi.13172.
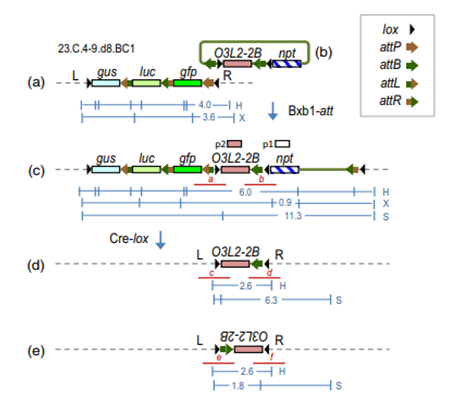
Figure. Schematic map of Bxb1 & Cre -mediated gene replacement.
